Hamming Distance and Hashing Functions
Description of the Problem
As yesterday, assume \((X,d)\) is a metric space of bitstrings of a fixed length together with the Hamming distance. Today, I am going to look at the distributions of numbers \(d(f(x),x)\) for \(x\in X\).
Implementation
As yesterday, I am going to use ironclad:
(require :ironclad)
NILand the same MD5 and SHA1 hashing functions:
(let ((hasher (ironclad:make-digest :md5)))
(defun md5-digest (x)
(coerce (ironclad:digest-sequence hasher x) 'list)))
MD5-DIGEST
(let ((hasher (ironclad:make-digest :sha1)))
(defun sha1-digest (x)
(coerce (ironclad:digest-sequence hasher x) 'list)))
SHA1-DIGESTSince the space of bit strings of length 128 is large, I will use a Monte-Carlo method:
(defun random-vector (m)
(coerce
(loop repeat m collect (random 256))
'(vector (unsigned-byte 8))))
RANDOM-VECTORAnd finally, the function that calculates the quantity I am interested in is
(defun hamming-distance (x y)
(labels ((calc (x y &optional (acc 0))
(cond
((< x y) (calc y x 0))
((zerop x) acc)
(t (calc (ash x -1)
(ash y -1)
(if (equal (oddp x) (oddp y))
acc
(1+ acc)))))))
(reduce #'+ (map 'list (lambda (i j) (calc i j)) x y))))
HAMMING-DISTANCENow, let me test this on a sample of size 40,000 on MD5:
(with-open-file (out "hamming2-data1.csv" :direction :output
:if-exists :supersede
:if-does-not-exist :create)
(loop repeat 40000 do
(let ((x (random-vector 16)))
(format out "~4,9f~%" (/ (hamming-distance (md5-digest x) x) 1.0)))))
NILHere is a histogram of the data:
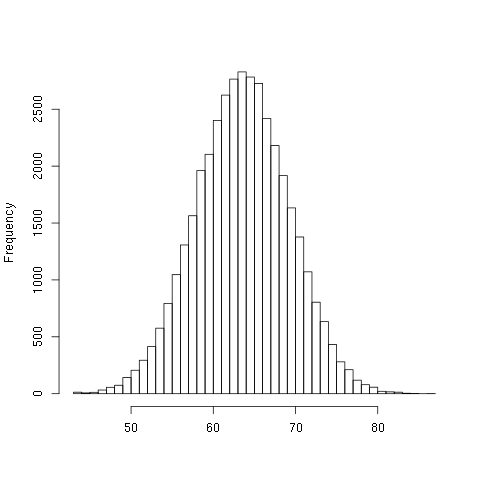
For the majority of the vectors, it is most likely that about 50% of the bits of the output will be different than the input.
Now, let me test this again on SHA1. Recall that since SHA1 digest produces 20 bytes, I am going to need random vectors of length 20:
(with-open-file (out "hamming2-data2.csv" :direction :output
:if-exists :supersede
:if-does-not-exist :create)
(loop repeat 40000 do
(let ((x (random-vector 20)))
(format out "~4,9f~%" (/ (hamming-distance (sha1-digest x) x) 1.0)))))
NILHere is a histogram of the data:
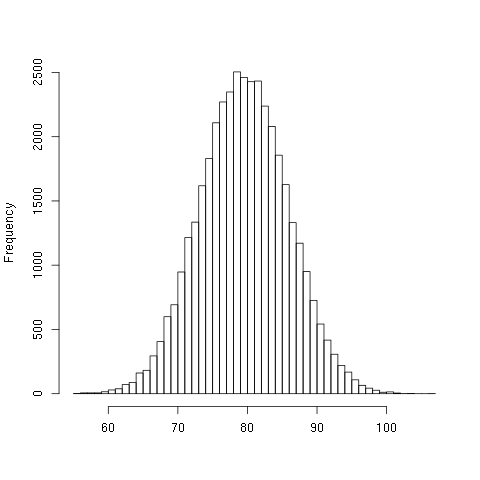
Again, about 50% of the bits are different between the input and output vectors.
An Analysis
I took the data from these experiments and normalized the values to the interval [0,1]. When I compared the distributions with the Kolmogorov-Smirnov test I see that
Two-sample Kolmogorov-Smirnov test
data: normalize(as.matrix(X)) and normalize(as.matrix(Y))
D = 0.2524, p-value < 2.2e-16
alternative hypothesis: two-sidedwhich means that the distributions are different. Very interesting!